OnnxPredictionSegmentation2d
Computes a prediction on a two-dimensional image from an ONNX model and applies a post processing to generate a label or a binary image.
Access to parameter description
For an introduction, please refer to the Deep Learning section.
The following steps are applied:
If the prediction output contains two channels or more, the channel index presenting the highest score is assigned to each pixel of the output image. The first class, represented in the channel of index 0, is considered as background. The output image has the label interpretation in this case.
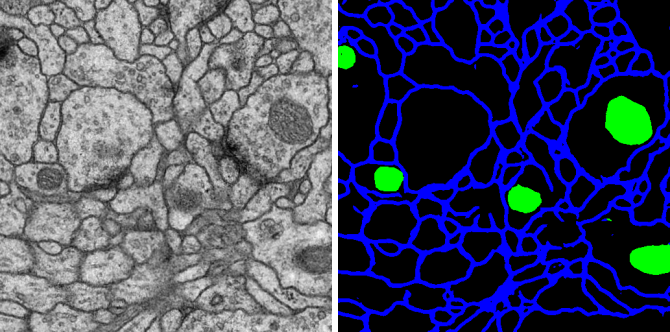
Figure 1. Membrane segmentation by deep learning prediction with a Unet model trained with the Avizo software.
This image is shown by courtesy of Albert Cardona. It is from a set of serial section TEM images of Drosophila brain tissue described in this paper: Cardona A, et al. 2010. An Integrated Micro- and Macroarchitectural Analysis of the Drosophila Brain by Computer-Assisted Serial Section Electron Microscopy. PLoS Biol 8(10): e1000502. doi:10.1371/journal.pbio.1000502.
See also
Access to parameter description
For an introduction, please refer to the Deep Learning section.
The following steps are applied:
- Pre-processing
- Prediction
- Post-processing to generate a binary or label output
If the prediction output contains two channels or more, the channel index presenting the highest score is assigned to each pixel of the output image. The first class, represented in the channel of index 0, is considered as background. The output image has the label interpretation in this case.

Figure 1. Membrane segmentation by deep learning prediction with a Unet model trained with the Avizo software.
This image is shown by courtesy of Albert Cardona. It is from a set of serial section TEM images of Drosophila brain tissue described in this paper: Cardona A, et al. 2010. An Integrated Micro- and Macroarchitectural Analysis of the Drosophila Brain by Computer-Assisted Serial Section Electron Microscopy. PLoS Biol 8(10): e1000502. doi:10.1371/journal.pbio.1000502.
See also
Function Syntax
This function returns outputObjectImage.
// Function prototype
std::shared_ptr< iolink::ImageView > onnxPredictionSegmentation2d( std::shared_ptr< iolink::ImageView > inputImage, std::string modelPath, OnnxPredictionSegmentation2d::DataFormat dataFormat, OnnxPredictionSegmentation2d::InputNormalizationType inputNormalizationType, iolink::Vector2d normalizationRange, OnnxPredictionSegmentation2d::NormalizationScope normalizationScope, iolink::Vector2u32 tileSize, uint32_t tileOverlap, std::shared_ptr< iolink::ImageView > outputObjectImage = NULL );
This function returns outputObjectImage.
// Function prototype.
onnx_prediction_segmentation_2d( input_image,
model_path = "",
data_format = OnnxPredictionSegmentation2d.DataFormat.NHWC,
input_normalization_type = OnnxPredictionSegmentation2d.InputNormalizationType.STANDARDIZATION,
normalization_range = [0, 1],
normalization_scope = OnnxPredictionSegmentation2d.NormalizationScope.GLOBAL,
tile_size = [256, 256],
tile_overlap = 32,
output_object_image = None )
This function returns outputObjectImage.
// Function prototype.
public static IOLink.ImageView
OnnxPredictionSegmentation2d( IOLink.ImageView inputImage,
String modelPath = "",
OnnxPredictionSegmentation2d.DataFormat dataFormat = ImageDev.OnnxPredictionSegmentation2d.DataFormat.NHWC,
OnnxPredictionSegmentation2d.InputNormalizationType inputNormalizationType = ImageDev.OnnxPredictionSegmentation2d.InputNormalizationType.STANDARDIZATION,
double[] normalizationRange = null,
OnnxPredictionSegmentation2d.NormalizationScope normalizationScope = ImageDev.OnnxPredictionSegmentation2d.NormalizationScope.GLOBAL,
uint[] tileSize = null,
UInt32 tileOverlap = 32,
IOLink.ImageView outputObjectImage = null );
Class Syntax
Parameters
| Class Name | OnnxPredictionSegmentation2d |
|---|
| Parameter Name | Description | Type | Supported Values | Default Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 |
inputImage |
The input image. It can be a grayscale or color image, depending on the selected model. | Image | Binary, Label, Grayscale or Multispectral | nullptr | ||||||
 |
modelPath |
The path to the ONNX model file. | String | "" | |||||||
 |
dataFormat |
The tensor layout expected as input by the model. The input image is automatically converted to this layout by the algorithm.
|
Enumeration | NHWC | |||||||
 |
inputNormalizationType |
The type of normalization to apply before computing the prediction. It is recommended to apply the same pre-processing as during the training.
|
Enumeration | STANDARDIZATION | |||||||
 |
normalizationRange |
The data range in which the input image is normalized before computing the prediction. It is recommended to apply the same pre-processing as during the training. This parameter is ignored if the normalization type is set to none. | Vector2d | Any value | {0.f, 1.f} | ||||||
 |
normalizationScope |
The scope for computing normalization (mean, standard deviation, minimum or maximum). This parameter is ignored if the normalization type is set to none.
|
Enumeration | GLOBAL | |||||||
 |
tileSize |
The width and height in pixels of the sliding window. This size includes the user defined tile overlap. | Vector2u32 | Any value | {256, 256} | ||||||
 |
tileOverlap |
The number of pixels used as overlap between the tiles. An overlap of zero may lead to artifacts in the prediction result. A non-zero overlap reduces such artifacts but increases the computation time. | UInt32 | Any value | 32 | ||||||
 |
outputObjectImage |
The output image. Dimensions, and calibration of the output image are forced to the same values as the input. Its interpretation is binary if the model produces one channel, label otherwise. | Image | nullptr | |||||||
Object Examples
std::shared_ptr< iolink::ImageView > membrane = ioformat::readImage( std::string( IMAGEDEVDATA_IMAGES_FOLDER ) + "membrane.png" );
OnnxPredictionSegmentation2d onnxPredictionSegmentation2dAlgo;
onnxPredictionSegmentation2dAlgo.setInputImage( membrane );
onnxPredictionSegmentation2dAlgo.setModelPath( std::string( IMAGEDEVDATA_OBJECTS_FOLDER ) + "membrane.onnx" );
onnxPredictionSegmentation2dAlgo.setDataFormat( OnnxPredictionSegmentation2d::DataFormat::NHWC );
onnxPredictionSegmentation2dAlgo.setInputNormalizationType( OnnxPredictionSegmentation2d::InputNormalizationType::STANDARDIZATION );
onnxPredictionSegmentation2dAlgo.setNormalizationRange( {0, 1} );
onnxPredictionSegmentation2dAlgo.setNormalizationScope( OnnxPredictionSegmentation2d::NormalizationScope::GLOBAL );
onnxPredictionSegmentation2dAlgo.setTileSize( {128, 128} );
onnxPredictionSegmentation2dAlgo.setTileOverlap( 32 );
onnxPredictionSegmentation2dAlgo.execute();
std::cout << "outputObjectImage:" << onnxPredictionSegmentation2dAlgo.outputObjectImage()->toString();
membrane = ioformat.read_image(imagedev_data.get_image_path("membrane.png"))
onnx_prediction_segmentation_2d_algo = imagedev.OnnxPredictionSegmentation2d()
onnx_prediction_segmentation_2d_algo.input_image = membrane
onnx_prediction_segmentation_2d_algo.model_path = imagedev_data.get_object_path("membrane.onnx")
onnx_prediction_segmentation_2d_algo.data_format = imagedev.OnnxPredictionSegmentation2d.NHWC
onnx_prediction_segmentation_2d_algo.input_normalization_type = imagedev.OnnxPredictionSegmentation2d.STANDARDIZATION
onnx_prediction_segmentation_2d_algo.normalization_range = [0, 1]
onnx_prediction_segmentation_2d_algo.normalization_scope = imagedev.OnnxPredictionSegmentation2d.GLOBAL
onnx_prediction_segmentation_2d_algo.tile_size = [128, 128]
onnx_prediction_segmentation_2d_algo.tile_overlap = 32
onnx_prediction_segmentation_2d_algo.execute()
print( "output_object_image:", str( onnx_prediction_segmentation_2d_algo.output_object_image ) )
ImageView membrane = ViewIO.ReadImage( @"Data/images/membrane.png" );
OnnxPredictionSegmentation2d onnxPredictionSegmentation2dAlgo = new OnnxPredictionSegmentation2d
{
inputImage = membrane,
modelPath = @"Data/objects/membrane.onnx",
dataFormat = OnnxPredictionSegmentation2d.DataFormat.NHWC,
inputNormalizationType = OnnxPredictionSegmentation2d.InputNormalizationType.STANDARDIZATION,
normalizationRange = new double[]{0, 1},
normalizationScope = OnnxPredictionSegmentation2d.NormalizationScope.GLOBAL,
tileSize = new uint[]{128, 128},
tileOverlap = 32
};
onnxPredictionSegmentation2dAlgo.Execute();
Console.WriteLine( "outputObjectImage:" + onnxPredictionSegmentation2dAlgo.outputObjectImage.ToString() );
Function Examples
std::shared_ptr< iolink::ImageView > membrane = ioformat::readImage( std::string( IMAGEDEVDATA_IMAGES_FOLDER ) + "membrane.png" );
auto result = onnxPredictionSegmentation2d( membrane, std::string( IMAGEDEVDATA_OBJECTS_FOLDER ) + "membrane.onnx", OnnxPredictionSegmentation2d::DataFormat::NHWC, OnnxPredictionSegmentation2d::InputNormalizationType::STANDARDIZATION, {0, 1}, OnnxPredictionSegmentation2d::NormalizationScope::GLOBAL, {128, 128}, 32 );
std::cout << "outputObjectImage:" << result->toString();
membrane = ioformat.read_image(imagedev_data.get_image_path("membrane.png"))
result = imagedev.onnx_prediction_segmentation_2d( membrane, imagedev_data.get_object_path("membrane.onnx"), imagedev.OnnxPredictionSegmentation2d.NHWC, imagedev.OnnxPredictionSegmentation2d.STANDARDIZATION, [0, 1], imagedev.OnnxPredictionSegmentation2d.GLOBAL, [128, 128], 32 )
print( "output_object_image:", str( result ) )
ImageView membrane = ViewIO.ReadImage( @"Data/images/membrane.png" );
IOLink.ImageView result = Processing.OnnxPredictionSegmentation2d( membrane, @"Data/objects/membrane.onnx", OnnxPredictionSegmentation2d.DataFormat.NHWC, OnnxPredictionSegmentation2d.InputNormalizationType.STANDARDIZATION, new double[]{0, 1}, OnnxPredictionSegmentation2d.NormalizationScope.GLOBAL, new uint[]{128, 128}, 32 );
Console.WriteLine( "outputObjectImage:" + result.ToString() );